- KOR
- |
- ENG
eGnome's Publications
CNS paper list (total 6 papers)
eGnome's Publications
Selected as Proud Koreans(Hanbit-Sa) paper list (total 12 papers)
eGnome's Publications
paper list (total 154 papers)
-
2022~Current
1. Genomic insights and functional evaluation of Lacticaseibacillus paracasei EG005: a promising probiotic with enhanced antioxidant activity. Frontiers in Microbiology (2024.10)
2. Identifying genes within pathways in unannotated genomes with PaGeSearch. Genome Research (2024.06)
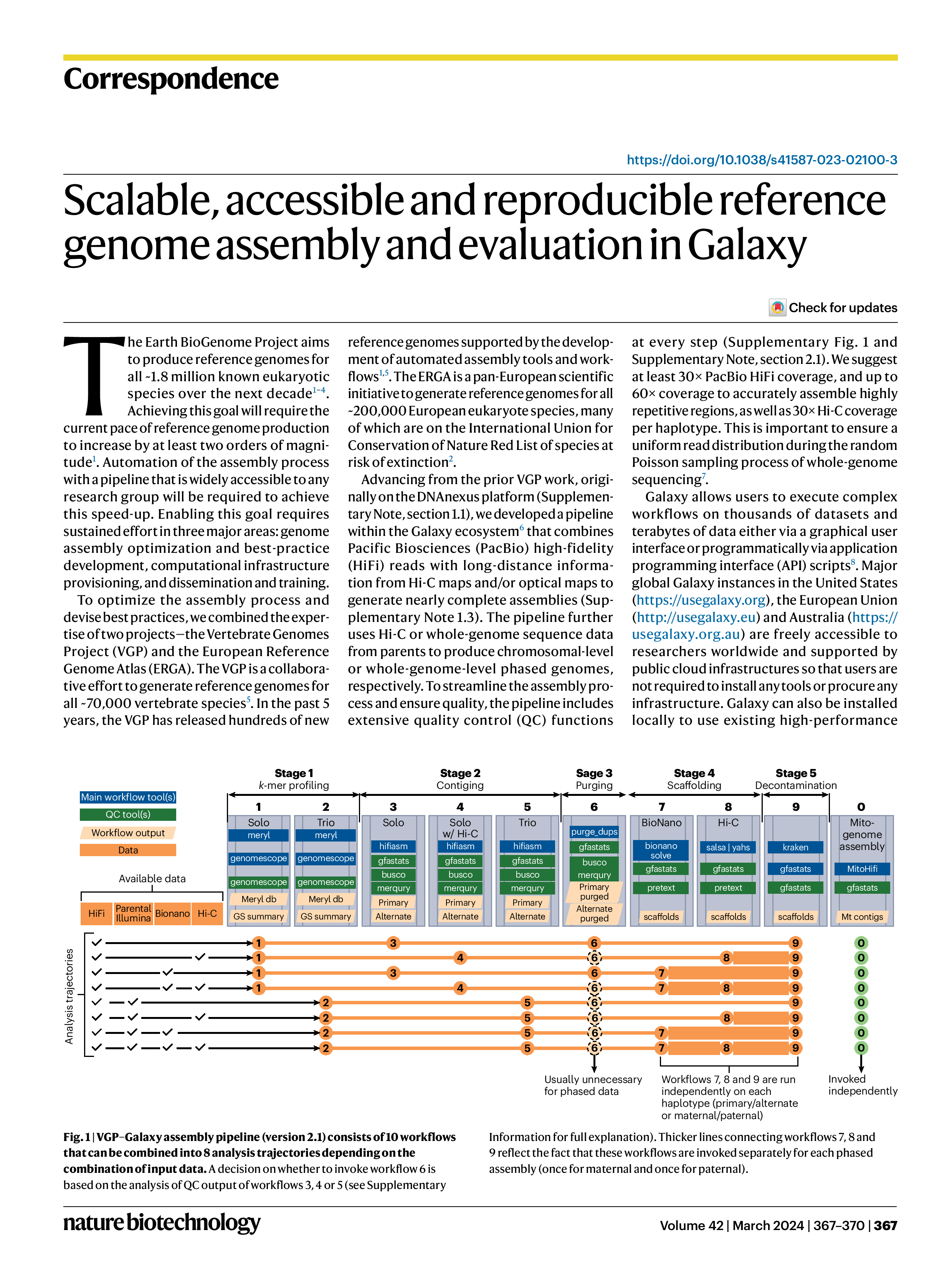
3. Scalable, accessible and reproducible reference genome assembly and evaluation in Galaxy. Nature Biotechnology (2024.01)
4. Chromosome-level genome assembly of chub mackerel (Scomber japonicus) from the Indo-Pacific Ocean. Scientific Data (2023.12)
5. Inference of Admixture Origins in Indigenous African Cattle. Molecular Biology and Evolution (2023.11)
6. Chromosome-level genome assembly of Korean native cattle and pangenome graph of 14 Bos taurus assemblies. Scientific Data (2023.08)
7. HGTree v2.0: a comprehensive database update for horizontal gene transfer (HGT) events detected by the tree-reconciliation method. Nucleic Acids Research (2022.11)
8. Widespread false gene gains caused by duplication errors in genome assemblies. Genome Biology (2022.09)
9. Investigation of memory-enhancing effects of Streptococcus thermophilus EG007 in mice and elucidating molecular and metagenomic characteristics using Nanopore sequencing. Scientific Reports (2022.08)
10. In silico SARS-CoV-2 vaccine development for Omicron strain using reverse vaccinology. Genes & Genomics (2022.06)
11. Shared genetic etiology and antagonistic relationship of plasma renin activity and systolic blood pressure in a Korean cohorts. Genomics (2022.05)
12. Microbial Identification Using rRNA Operon Region: Database and Tool for Metataxonomics with Long-Read Sequence. Microbiology spectrum (2022.03)
13. Immunogenic Epitope-Based Vaccine Prediction from Surface Glycoprotein of MERS-CoV by Deploying Immunoinformatics Approach. International Journal of Peptide Research and Therapeutics (2022.03)
14. Positive Effect of Lactobacillus acidophilus EG004 on Cognitive Ability of Healthy Mice by Fecal Microbiome Analysis Using Full-Length 16S-23S rRNA Metagenome Sequencing. Microbiology spectrum (2022.01)
-
2017~2021
1. Complete Genome Sequence of the Newly Developed Lactobacillus acidophilus Strain With Improved Thermal Adaptability. Frontiers in Microbiology (2021.09)
2. Population differentiated copy number variation of Bos taurus, Bos indicus and their African hybrids. BMC GENOMICS (2021.07)
3. Selection and evaluation of bi-allelic autosomal SNP markers for paternity testing in Koreans. INTERNATIONAL JOURNAL OF LEGAL MEDICINE (2021.04)
4. Towards complete and error-free genome assemblies of all vertebrate species. Nature (2021.04)
5. False gene and chromosome losses in genome assemblies caused by GC content variation and repeats. Genome biology (2021.04)
6. Widespread false gene gains caused by duplication errors in genome assemblies. Genome biology (2021.04)
7. False gene and chromosome losses affected by assembly and sequence errors. bioRxiv (2021.04)
8. Genotyping-by-Sequencing of the regional Pacific abalone (Haliotis discus) genomes reveals population structures and patterns of gene flow. PLOS ONE (2021.04)
9. Genetic Adaptations in Mudskipper and Tetrapod Give Insights into Their Convergent Water-to-Land Transition. Animals (2021.02)
10. Mitonuclear incompatibility as a hidden driver behind the genome ancestry of African admixed cattle. BMC Biology (2021.01)
11. The mosaic genome of indigenous African cattle as a unique genetic resource for African pastoralism. Nature Genetics (2020.09)
12. Complete Genomic Analysis of Enterococcus faecium Heat-Resistant Strain Developed by Two-Step Adaptation Laboratory Evolution Method. Front. Bioeng. Biotechnol. (2020.07)
13. Is LactobacillusGram-Positive? A Case Study ofLactobacillus iners. Microorganisms (2020.06)
14. Genome-Wide Identification of Discriminative Genetic Variations in Beef and Dairy Cattle via an Information-Theoretic Approach. Genes (2020.06)
15. Changes in Cell Membrane Fatty Acid Composition of Streptococcus thermophilus in Response to Gradually Increasing Heat Temperature. J. Microbiol Biotechnol. (2020.05)
16. Complete Genome ofLactobacillus inersKY Using Flongle Provides Insight Into the Genetic Background of Optimal Adaption to Vaginal Econiche. Front. Microbiol. (2020.05)
17. Genomic Prediction Accuracy Using Haplotypes Defined by Size and Hierarchical Clustering Based on Linkage Disequilibrium. Front Genet. (2020.03)
18. Identification of Copy Number Variation in Domestic Chicken Using Whole-Genome Sequencing Reveals Evidence of Selection in the Genome. Animals (2019.10)
19. Expression and Purification of Extracellular Solute-Binding Protein (ESBP) in Escherichia coli, the Extracellular Protein Derived from Bifidobacterium longum KACC 91563. Food Sci. Anim. Resour. (2019.08)
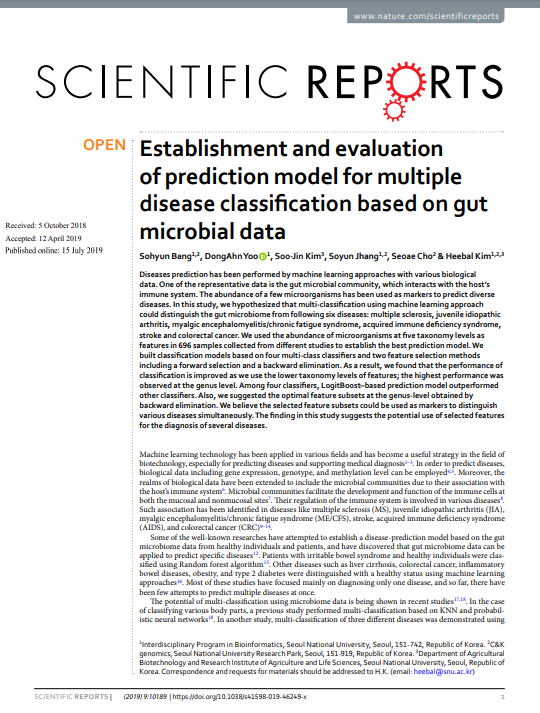
20. Establishment and evaluation of prediction model for multiple disease classification based on gut microbial data. Scientific Reports (2019.07)
21. Bayesian evolutionary hypernetworks for interpretable learning from high-dimensional data. Applied Soft Computing (2019.05)
22. De novo emergence and potential function of human-specific tandem repeats in brain-related loci. Human Genetics (2019.05)
23. Whole genome sequencing reveals the impact of recent artificial selection on red sea bream reared in fish farms. Scientific Reports (2019.04)
24. Exploring the genomes of East African Indicine cattle breeds reveals signature of selection for tropical environmental adaptation traits. Cogent Food Agr. (2018.11)
25. Deciphering the evolutionary signatures of pinnipeds using novel genome sequences: The first genomes of Phoca largha, Callorhinus ursinus, and Eumetopias jubatus. Scientific Reports (2018.11)
26. Zygotic gene activation in the chicken occurs in two waves, the first involving only maternally derived genes. eLife (2018.10)
27. Microbial community and functions associated with digestion of algal polysaccharides in the visceral tract of Haliotis discus hannai: Insights from metagenome and metatranscriptome analysis. PLoS One (2018.10)
28. Rhizoshere microbiome structure alters to enable wilt resistance in tomato. Nature Biotechnology (2018.10)
29. Genome sequencing and protein domain annotations of Korean Hanwoo cattle identify Hanwoo-specific immunity-related and other novel genes. BMC Genetics (2018.05)
30. Horizontal gene transfer of Chlamydia: Novel insights from tree reconciliation. PLoS ONE (2018.04)
31. The first whole transcriptomic exploration of pre-oviposited early chicken embryos using single and bulked embryonic RNA-sequencing. GigaScience (2018.04)
32. Coelacanth-specific adaptive genes give insights into primitive evolution for water-to-land transition of tetrapods. Marine Genomics (2018.04)
33. Artificial selection increased body weight but induced increase of runs of homozygosity in Hanwoo cattle. PLoS One (2018.03)
34. The transcriptome of early chicken embryos reveals signaling pathways governing rapid asymmetric cellularization and lineage segregation. Development (2018.03)
35. Identification of genes related to intramuscular fat content of pigs using genome-wide association study. Asian Australas. J. Anim. Sci. (2018.02)
36. Transcriptional and translational dynamics during maternal-to-zygotic transition in early chicken development. FASEB journal (2018.01)
37. Comparative genomic analysis of Lactobacillus plantarum GB-LP4 and identification of evolutionarily divergent genes in high-osmolarity environment. Genes & Genomics (2017.11)
38. De novo-based transcriptome profiling of male-sterile and fertile watermelon lines. PLoS ONE (2017.11)
39. Genetic and phenotypic characterization of the novel mouse substrain C57BL/6N Korl with increased body weight. Scientific Reports (2017.10)
40. Deciphering signature of selection affecting beef quality traits in Angus cattle. Genes & Genomics (2017.09)
41. Methanobrevibacter Attenuation via Probiotic Intervention Reduces Flatulence in Adult Human: A Non-Randomised Paired-Design Clinical Trial of Efficacy. PLoS ONE (2017.09)
42. Exploring evidence of positive selection signatures in cattle breeds selected for different traits . Mammalian Genome (2017.09)
43. The Genetic Origin of Short Tail in Endangered Korean Dog, DongGyeongi. Scientific Reports (2017.08)
44. Complete Genome Analysis of Lactobacillus fermentum SK152 from Kimchi Reveals Genes Associated with its Antimicrobial Activity. FEMS Microbiology Letters (2017.08)
45. Whole genome detection of signature of positive selection in African cattle reveals selection for thermotolerance. Animal Science Journal (2017.07)
46. Comparative genome analysis of Lactobacillus plantarum GB-LP1 isolated from traditional Korean fermented food . Journal of Microbiology and Biotechnology (2017.06)
47. Comparative genome analysis of Lactobacillus plantarum GB-LP3 provides candidates of survival-related genetic factors. Infection, Genetics and Evolution (2017.05)
48. Comparative genomic analysis reveals genetic features related to the virulence of Bacillus cereus FORC_013. Gut Pathogens (2017.05)
49. Cattle genome-wide analysis reveals genetic signatures in trypanotolerant N’Dama. BMC Genomics (2017.05)
50. Identifying candidate positive selection genes in Korean imported pig breeds. Genes & Genomics (2017.05)
51. Complete mitochondrial genome sequences of Korean native horse from Jeju Island: Uncovering the spatio-temporal dynamics.. Molecular Biology Reports (2017.04)
52. Genome sequence of pacific abalone (Haliotis discus hannai): the first draft genome in family Haliotidae.. Gigascience (2017.03)
53. Genome-association analysis of Korean Holstein milk traits using genomic estimated breeding value. Asian-Austraiasian Journal of Animal Sciences (2017.03)
54. Genomic Insights and Its Comparative Analysis with Yersinia enterocolitica Reveals the Potential Virulence Determinants and Further Pathogenicity for Foodborne Outbreaks.. J. Microbiol. Biotechnol (2017.02)
55. The genome landscape of indigenous African cattle. Genome Biology (2017.02)
56. Whole genome scan reveals the genetic signature of African Ankole cattle breed and potential for higher quality beef. BMC Genetics (2017.02)
57. Identification of a Novel Mutation in BRD4 that Causes Autosomal Dominant Syndromic Congenital Cataracts Associated with Other Neuro-Skeletal Anomalies. PLoS ONE (2017.01)
-
2012~2016
1. Detecting Positive Selection of Korean Native Goat Populations Using Next-Generation Sequencing. Molecules and Cells (2016.12)
2. Complete genome of Vibrio parahaemolyticus FORC014 isolated from the toothfish. Gut Pathogens (2016.11)
3. Gut microbiota Modulated by Probiotics and Garcinia cambogia Extract Correlate with Weight Gain and Adipocyte Sizes in High Fat-Fed Mice.. Scientific Reports (2016.09)
4. Engineering Structures and Functions of Mesenchymal Stem Cells by Suspended Large-Area Graphene Nanopatterns.. 2D Materials (2016.08)
5. Status of dosage compensation of X chromosome in bovine genome.. Genetica (2016.07)
6. Whole genome sequencing of Gyeongbuk Araucana, a newly developed blue-egg laying chicken breed, reveals its origin and genetic characteristics. Scientific Reports (2016.05)
7. Complete genome sequence of Vibrio parahaemolyticusstrain FORC_008, a foodborne pathogen from a flounder fish in South Korea. Pathogens and Disease (2016.05)
8. Evolutionary constraints over microsatellite abundance in larger mammals as a potential mechanism against carcinogenic burden. Scientific Reports (2016.04)
9. Comparative analysis of the complete genome of Lactobacillus Plantarum GB-LP2 and potential candidate genes for host immune system enhancement. Journal of Microbiology and Biotechnology (2016.04)
10. Effects of road transportation on metabolic and immunological responses in Holstein heifers. Animal Science Journal (2016.04)
11. RNA-seq analysis for detecting quantitative trait-associated genes. Scientific Reports (2016.04)
12. Complete genome sequence of Vibrio parahaemolyticus FORC_023 isolated from raw fish storage water.. Pathogens and Disease (2016.04)
13. Characterizing milk production related genes in Holstein using RNA-seq. Asian-Australasian Journal of Animal Sciences (2016.02)
14. Comprehensive identification of sexually dimorphic genes in diverse cattle tissues using RNA-seq.. BMC Genomics (2016.01)
15. A Genome-wide Scan for Selective Sweeps in Racing Horses. Asian-Australasian Journal of Animal Sciences (2015.11)
16. Analysis of Stage-Specific Gene Expression Profiles in the Uterine Endometrium during Pregnancy in Pigs. . PLoS ONE (2015.11)
17. HGTree: database of horizontally transferred genes determined by tree reconciliation.. Nucleic Acids Research (2015.11)
18. Transcriptome profiling of differentially expressed genes in floral buds and flowers of male sterile and fertile lines in watermelon.. BMC Genomics (2015.11)
19. Genome-wide association study (GWAS) and its application for improving the genomic estimated breeding values (GEBV) of the Berkshire pork quality traits. Asian-Australasian Journal of Animal Sciences (2015.11)
20. A novel splice variant of the decapentaplegic (dpp) gene in the wild silkworm, Bombyx mandarina. Biochemical and Biophysical Research Communications (2015.10)
21. Application of LogitBoost Classifier for Traceability Using SNP Chip Data. PLoS ONE (2015.10)
22. Caloric restriction improves diabetes-induced cognitive deficits by attenuating neurogranin-associated calcium signaling in high-fat diet-fed mice. Journal of Cerebral Blood Flow & Metabolism (2015.10)
23. Comprehensive Identification of Sexual Dimorphism-Associated Differentially Expressed Genes in Two-Way Factorial Designed RNA-Seq Data on Japanese Quail (Coturnix coturnix japonica). PLoS ONE (2015.09)
24. The prediction of the expected current selection coefficient of single nucleotide polymorphism (SNP) associated with Holstein milk yield, fat and protein contents. Asian-Austraiasian Journal of Animal Sciences (2015.09)
25. Exploring evidence of positive selection reveals genetic basis of meat quality traits in Berkshire pigs through whole genome sequencing. BMC Genetics (2015.08)
26. Single high-dose irradiation aggravates eosinophil-mediated fibrosis through IL-33 secreted from impaired vessels in the skin compared to fractionated irradiation . Biochemical and Biophysical Research Communications (2015.08)
27. African Indigenous Cattle: Unique Genetic Resources in a Rapidly Changing World.. Asian-Australasian Journal of Animal Sciences (2015.07)
28. Complete genome sequence and SNPs of Raja pulchra (Rajiformes, Rajidae) mitochondria . Mitochondrial DNA (2015.06)
29. Developmental Dynamic Analysis of the Excreted Microbiome of Chickens Using Next-Generation Sequencing. Journal of Molecular Microbiology and Biotechnology (2015.06)
30. Multiple Genes Related to Muscle Identified through a Joint Analysis of a Two-stage Genome-wide Association Study for Racing Performance of 1, 156 Thoroughb reds.. Asian-Australasian Journal of Animal Sciences (2015.06)
31. Exploring the Genetic Signature of Body Size in Yucatan Miniature Pig. PLoS ONE (2015.04)
32. Genome Wide Association Studies Using Multiple-lactation Breeding Value in Holsteins. Asian-Australasian Journal of Animal Sciences (2015.03)
33. Time-calibrated phylogenomics of the classical swine fever viruses: genome-wide Bayesian . PLoS ONE (2015.03)
34. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project.. Genome Biology (2015.03)
35. Bovine Genome-wide Association Study for Genetic Elements to Resist the Infection of Foot-and-mouth Disease in the Field. Asian-Austraiasian Journal of Animal Sciences (2015.02)
36. A genome-wide scan for signatures of directional selection in domesticated pigs. BMC Genomics (2015.02)
37. Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage. BMC Plant Biology (2015.02)
38. Genome-wide detection and characterization of positive selection in Korean Native Black Pig from Jeju Island.. BMC Genetics (2015.01)
39. Cetaceans evolution: insights from the genome sequences of common minke whales.. BMC Genomics (2015.01)
40. The usage of SNP-SNP relationship matrix in the best linear unbiased prediction (BLUP) analysis using the community based cohort study. Genomics & Informatics (2014.12)
41. Comparative genomics reveals insights into avian genome evolution and adaptation. Science (2014.12)
42. Uncovering genomic features and maternal origin of Korean Native Chicken by whole genome sequencing.. PLoS ONE (2014.12)
43. Genomic Selection for Adjacent Genetic Markers of Yorkshire Pigs Using Regularized Regression Approaches.. Asian-Australasian Journal of Animal Sciences (2014.12)
44. VCS: tool for Visualizing Copy number variation and Single nucleotide polymorphism.. Asian-Austraiasian Journal of Animal Sciences. (2014.12)
45. Genome-wide DNA Methylation Profiles of Small Intestine and Liver in Fast-growing and Slow-growing Weaning Piglets. Asian-Austraiasian Journal of Animal Sciences (2014.11)
46. Comparative Genomic Analysis of Staphylococcus aureus FORC_001 and S. aureus MRSA252 Reveal the Characteristics of Antibiotic Resistance and Virulence Factors for Human Infection. Journal of Microbiology and Biotechnology (2014.10)
47. The association of SLC6A4 5-HTTLPR and TRPV1 945G>C with functional dyspepsia in Korea.. Journal of Gastroenterology and Hepatology (2014.10)
48. A Novel Genetic Variant Database for Korean Native Cattle (Hanwoo): HanwooGDB. Genes & Genomics (2014.10)
49. Estimating effective population size of thoroughbred horses using linkage disequilibrium and theta (4Nμ) value.. Livestock Science (2014.10)
50. Copy Number Deletion Has Little Impact on Gene Expression Levels in Racehorses. Asian-Austraiasian Journal of Animal Sciences (2014.09)
51. Thoroughbred Horse Single Nucleotide Polymorphism and Expression Database: HSDB. Asian-Austraiasian Journal of Animal Sciences (2014.09)
52. Native Pig and Chicken Breed Database: NPCDB. Asian-Austraiasian Journal of Animal Sciences (2014.08)
53. Comparative transcriptomic analysis to identify differentially expressed genes in fat tissue of adult Berkshire and Jeju Native Pig using RNA-seq. Molecular Biology Reports (2014.07)
54. Gene expression profiling of bovine mammary gland epithelial cells stimulated with lipoteichoic acid plus peptidoglycan from Staphylococcus aureus. International Immunopharmacology (2014.07)
55. Genetic traceability of black pig meats using microsatellite markers. Asian-Austraiasian Journal of Animal Sciences (2014.07)
56. Deciphering the genetic blueprint behind Holstein milk proteins and production. Genome Biology and Evolution (2014.05)
57. Differential Evolution between Monotocous and Polytocous Species.. Asian-Australasian Journal of Animal Sciences (2014.04)
58. Semantic Modeling for SNPs associated with ethnic disparities in HapMap samples.. Genomics & Informatics (2014.03)
59. Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level.. BMC Genomics (2014.03)
60. Investigation of de novo unique differentially expressed genes related to evolution in exercise response during domestication in Thoroughbred race horses.. PLoS ONE (2014.03)
61. Genotype-environment interactions for quantitative traits in Korea Associated Resource (KARE) cohorts.. BMC Genetics (2014.02)
62. Characterization of Genes for Beef Marbling Based on Applying Gene Coexpression Network.. International Journal of Genomics (2014.01)
63. Genome-wide association study of integrated meat quality-related t raits of the Duroc pig breed.. Asian-Austraiasian Journal of Animal Sciences (2013.11)
64. Genome-wide Association Study of Chicken Plumage Pigmentation. . Asian-Australasian Journal of Animal Sciences (2013.10)
65. Genetic variants and signatures of selective sweep of Hanwoo population (Korean native cattle).. BMB Reports (2013.07)
66. Identification of differentially evolved genes: An alternative approach to detection of accelerated molecular evolution from genome-wide comparative data.. Evolutionary Bioinformatics (2013.07)
67. De novo assembly and comparative analysis of the Enterococcus faecalis genome (KACC91532) from a Korean neonate.. Journal of Microbiology and Biotechnology (2013.07)
68. Comparative transcriptome analysis of adipose tissues reveals that ECM-receptor interaction involved in the depot-specific adipogenesis in cattle.. PLoS One (2013.07)
69. Semantic networks for genome-wide CNV associated with AST and ALT in Korean cohorts.. Molecular & Cellular Toxicology (2013.07)
70. Acceleration of X-chromosome gene order evolution in the cattle lineage.. BMB Reports (2013.06)
71. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species.. Nature Genetics (2013.06)
72. Metagenome Analysis of Protein Domain Collocation within Cellulase Genes of Goat Rumen Microbes.. Asian-Austraiasian Journal of Animal Sciences (2013.05)
73. Peeling Back the Evolutionary Layers of Molecular Mechanisms Responsive to Exercise-Stress in the Skeletal Muscle of the Racing Horse.. DNA Research (2013.04)
74. Genetic diversity, population structure and relationships in indigenous cattle populations of Ethiopia and Hanwoo breeds using SNP markers.. Frontiers in Genetics (2013.03)
75. Ubiquitous polygenicity of human complex traits: genome-wide analysis of 49 traits in Koreans.. PLoS Genetics (2013.03)
76. Primed pluripotent cell lines derived from various embryonic origins and somatic cells in pig.. PLoS ONE (2013.01)
77. An integrated approach of comparative genomics and heritability analysis of pig and human on obesity trait: evidence for candidate genes on human chromosome 2.. BMC Genomics (2012.12)
78. Analyses of pig genomes provide insight into porcine demography and evolution.. Nature (2012.11)
79. Genome-wide analysis of copy number variations reveals that ageing processes influence body fat distribution in Korea Associated Resource (KARE) cohorts.. Human Genetics (2012.07)
80. Tracing the genetic history of porcine reproductive and respiratory syndrome viruses derived from the complete ORF 5-7 sequences: a Bayesian coalescent approach.. Archives of Virology (2012.07)
81. Gene expression profile of human peripheral blood mononuclear cells induced by Staphylococcus aureus lipoteichoic acid.. International Immunopharmacology (2012.05)
82. Microarray Analysis of Gene Expression in the Uterine Endometrium during the Implantation Period in Pigs.. Asian-Australasian Journal of Animal Sciences (2012.04)
83. Genome scanning for conditionally essential genes in Salmonella enterica Serotype Typhimurium.. Applied and Environmental Microbiology (2012.02)