
- KOR
- |
- ENG
이지놈에 오신 것을 환영합니다. 우리는 최첨단 3세대 마이크로바이옴 분석 기술을 자랑스럽게 소개합니다. 우리는 어려운 기술을 여러분이 이해하기 쉽도록 마이크로바이옴 분석을 슈퍼마켓에 비유하여 설명 드리겠습니다.
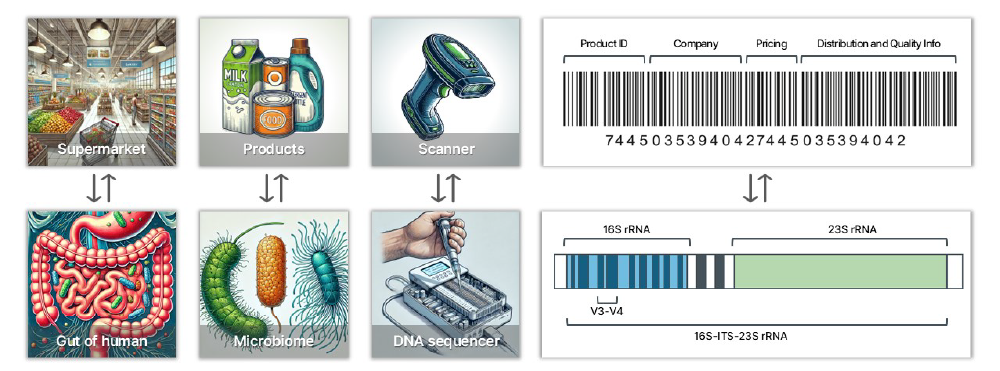
여러분이 슈퍼마켓에서 제공되는 제품의 종류와 수량을 확인하고자 한다고 가정해 봅시다. 각 제품의 바코드를 스캔하면 제조 회사와 특정 제품을 식별할 수 있습니다. 이 방법은 각 회사의 제품 종류와 수량에 대한 종합적인 데이터를 수집할 수 있게 해 줍니다.
이 비유를 우리의 마이크로바이옴 분석에 적용하면, 각 미생물의 rRNA 오페론(바코드에 해당하는 부분)을 분석하여 장내 마이크로바이옴(슈퍼마켓의 제품과 유사)을 조사합니다. rRNA 오페론은 16S, ITS, 그리고 23S 영역으로 구성되어 있으며, 이러한 영역을 읽는 것은 바코드를 스캔하여 세부 정보를 수집하는 것과 유사합니다. 대부분의 마이크로바이옴 헬스케어 회사들은 2세대 시퀀싱을 사용하여 16S 영역의 일부만(약 430개의 염기쌍) 읽어서 속(genus)수준의 정보를 제공합니다. 반면 이지놈의 3세대 시퀀싱 방식은 16S-ITS-23S 오페론 전체를 읽어서 종(species) 수준의 정보를 제공합니다. 이 방식은 약 4,300개의 염기쌍의 데이터를 읽어 들이기 때문에 2세대 방법보다 10배 더 많은 정보를 제공합니다.
예를 들어, 프로바이오틱스인 Lactobacillus acidophilus를 살펴보겠습니다. 'Lactobacillus'는 속(genus)를 나타내고, 'acidophilus'는 종(species)을 나타냅니다. 우리의 슈퍼마켓 비유에서 '속'은 '회사 이름'에 해당하고, '종'은 '제품 이름'에 해당합니다. 하나의 속은 100여 개의 서로 다른 종을 포함하고 있습니다.

종 수준의 식별은 100배 더 높은 해상도를 제공하여 유익균과 유해균을 정확하게 구별할 수 있게 합니다. 일부에서는 2세대 방법으로 종을 식별할 수 있다고 주장하지만, 그 정확도는 약 25%에 불과하여 신뢰할 수 없습니다. 이지놈의 방법은 종 식별에서 99.9%의 정확도를 자랑합니다. 우리는 옥스포드 나노포어 테크놀로지의 3세대 시퀀서를 사용하여 종 수준의 해상도를 정확하게 달성합니다. 7년간의 연구 끝에 우리는 2022년에 EG-Mirror 기술을 완성하였고, 그 결과를 Microbiology Spectrum 저널에 발표했습니다. 2024년에 우리는 EG-Mirror 버전 2를 출시했으며, 이는 현재 상업적으로 가장 정확한 플랫폼으로 인정받고 있습니다.
이 첨단 기술을 활용하여 이지놈은 다양한 마이크로바이옴 서비스를 제공합니다. 현재 장내 마이크로바이옴 분석 (EG gut PRO), 질내 마이크로바이옴 분석(EG vaginal PRO), 반려동물 장내 마이크로바이옴 분석(EG pet Dog, Cat) 서비스를 제공하고 있습니다. 향후 서비스에는 피부 마이크로바이옴 분석, 어린이 성장 및 발달 마이크로바이옴 분석, 비만 마이크로바이옴 분석 및 시니어 마이크로바이옴 분석이 포함될 예정입니다.

